Network Imaging
All sensations and behaviors are encoded in dynamic activity patterns of neural networks. In other words, complex structures of many individual neurons responding to environmental features, visual or auditory stimuli,, reward or punishment, etc. These neural systems extend over 3D space and, in most cases, cross many cortical layers of the brain. With two-photon microscopy enabling neuroscientists to reach the deep regions of the brain (down to 850 µm), it is now possible to study neural networks at a high spatiotemporal resolution.
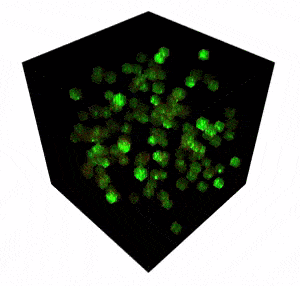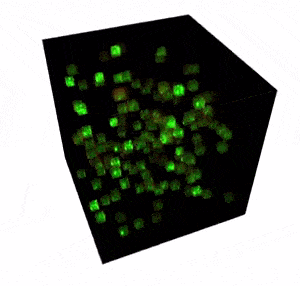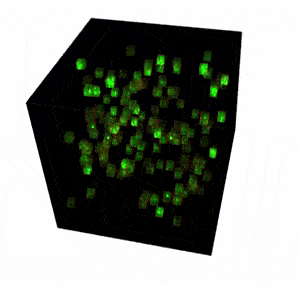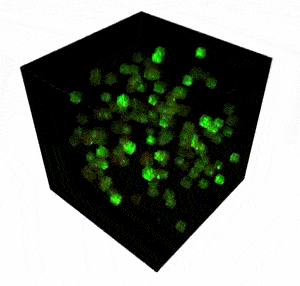


The FEMTO3D Atlas microscope with its unique technical solutions enables the precise spatial and real-time complexity of neuronal coding to be resolved by scanning large number of cells distributed in a 3D volume.
- Imaging of thousands of cells simultaneously
- Maximal scanning speed: 100 kHz
- High singal-to-noise ratio
- Online motion correction even in the axial dimension in behaving animal models
- Quasi-simultaneous 3D photostimulation with 3D imaging
- All available in the FEMTO3D Atlas two-photon imaging system
The imaging of cell populations in 3D is the fastest with FEMTO3D Atlas
3D random-access point scanning
Several thousand cells can be measured near-simultaneously (up to 100 kHz per point) by 3D random-access point scanning by restricting the imaging to sub-regions of the 3D volume, which results ultra-high scanning speed and signal-to-noise ratio.The figure (A-D) shows the changes in Ca2+ concentration of 2000 cells from the visual cortex of a GCaMP-expressing mouse as a function of time. See more:
Wertz et al., Science, 2015; Katona et al., Nat Meth, 2012. FEMTO3D Atlas

Imaging of multiple somata distributed in 3D in behaving animals
3D chessboard scanning
Chessboard scanning is a planar extension of random-access point scanning, where random-access points are extended to small squares by drifting the laser beam. These squares can be located anywhere in the imaging volume and include the somata with the surrounding area. Up to 300 somata can be measured simultaneously. The chessboard arrangement of the squares helps to visualize somata, analyze their activity, and correct for motion artifacts, and completely eliminating them with the help of the 3D Real-Time Motion Correction. The video shows in vivo neuronal activity, Ca2+ transients from 100 neuronal somata from the mouse V1 region, labeled with GCaMP6. See also Szalay et al., Neuron, 2016.
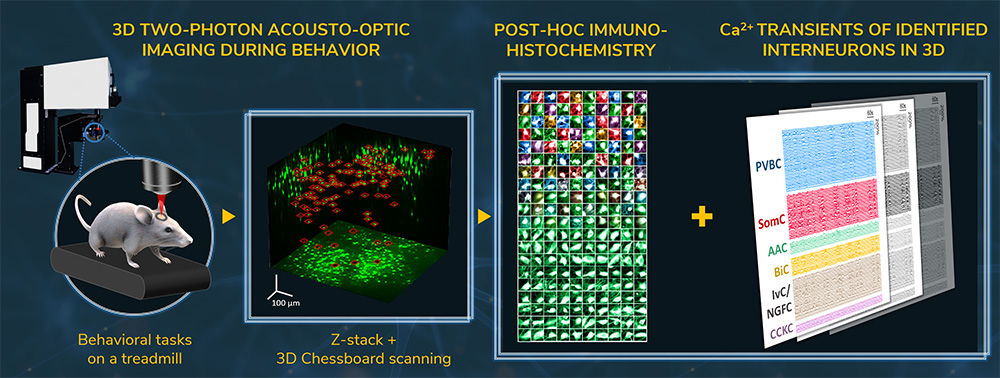
Imaging of sparsely distributed interneurons in a large neuronal network during behavior in real-time in 3D
3D chessboard scanning in a large neuronal network
Sparsely distributed brain cells (such as interneurons) can be followed in real-time in a 3D volume of using chessboard scanning. In the study depicted in the figure below, the calcium activity from nearly 250 interneurons was measured simultaneously, across every CA1 hippocampal layer, in an 800 × 800 × 500 µm volume with the chessboard scanning mode, while the mouse was performing behavioral tasks on a treadmill. Using these recordings, the biochemical composition of the imaged cells was also determined from data acquired from post hoc immunohistochemistry. Such a novelty – identifying distinct interneuron types and their activity dynamics simultaneously in a large neural network – could not have been achieved with other traditional recording techniques. See more: Geiller et al., Neuron, 2020. Watch the interview with one of the authors.
Imaging of multiple somata during large motions
3D multi-cube scanning
Multi-cube scanning is a spatially extended mode of chessboard scanning, where a Z dimension is added to the chessboard squares to cover the whole extent of the somata, therefore preserving all fluorescent information from the somata even during large amplitude movements Learn more: Bharioke et al. Neuron (2022)
Real-time motion-correction with the Femtonics 3D-RTMC
Our solution takes advantage of acousto-optics technology and elevates it to a whole new level. The flexible drift scanning method creates the opportunity to perform correction calculations with a predefined and adjustable frequency. The algorithm is based on correction calculations derived from selecting stable objects within the field of view (FOV), which serve as reference points during scanning. With our method providing an effortless means to scan in any direction in 3D, reference measurements are equally efficient in the Z direction as they are in the XY direction, which is a unique feature of the Femtonics 3D Real-Time Motion Correction.
Fastest frame scanning with rotational freedom
High-speed arbitrary frame scanning
Cells assembled into a neural network can be acquired with a frame rate of 40 Hz in a 500×500 µm field-of-view (FOV) by the High-speed arbitrary frame scanning mode of the Atlas. Beyond the high speed, another important and unique advantage of this mode is that the selected plane can be rotated into any in space to match your layer of interest.
The video below demonstrates the rotation of the selected plane in High-speed arbitrary frame scan mode. The left panel shows the relative position of the objective’s focal plane with blue and the actual two-photon focal plane with pink. The movement of the pink plane exemplifies the possibilities Atlas offers you to orient the plane in a three dimensional tissue. The right panel shows the neurons of a network in the actual scanned plane during rotation in live mode.

FEMTOSmart offers numerous imaging opportunities and with its vast modularity researchers can study neuronal networks from varied perspectives and save time in the experimental work.
Imaging of somata as ROIs in 2D
FEMTOSmart Galvo with flexible ROI scanning possibilities
The FEMTOSmart Galvo, with its flexible scanning patterns such as 2D random-access point, 2D multiple-line, and folded-frame scanning, supports the manual selection of individual cells in a neural network in a 2D plane. By skipping the measurement of the entire field, it is possible to reach a fast scanning speed and maintain a high signal-to-noise ratio. Using the TravellingSalesman additional software module, it is possible to determine the shortest pathway visiting defined points arbitrarily dispersed in the field of view. The short round-trip time results in a higher measurement repetition speed, up to 100 Hz for about 30 cells.
Multiple-line scanning
Fast scanning of the entire FOV or an entire volume
FEMTOSmart Resonant equipped with Piezo objective positioner kit
Fast-frame scanning by FEMTOSmart Resonant combined with fast Z-focusing performed by a Piezo objective positioner kit is a well-known approach for studying any three-dimensional neural network. In this case, the entire field of view is imaged continuously by the fast scanner, while the objective positioner moves between planes. More than one (cortical) layer might be recorded simultaneously this way, or the frames may be assembled into volumes resulting in a four-dimensional set of data.

References
Cortex-wide response mode of VIP-expressing inhibitory neurons by reward and punishment Zoltán Szadai, Hyun-Jae Pi, Quentin Chevy, Katalin Ócsai, Dinu F Albeanu, Balázs Chiovini, Gergely Szalay, Gergely Katona, Adam Kepecs (2022)
Large-Scale 3D Two-Photon Imaging of Molecularly Identified CA1 Interneuron Dynamics in Behaving Mice Tristan Geiller, Bert Vancura, Satoshi Terada, Eirini Troullinou, Spyridon Chavlis, Grigorios Tsagkatakis, Panagiotis Tsakalides, Katalin Ócsai, Panayiota Poirazi, Balázs J. Rózsa, Attila Losonczy / Neuron (2020)
Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution Cameron S. Cowan, Magdalena Renner, Martina De Gennaro, Brigitte Gross-Scherf, David Goldblum, Yanyan Hou, Martin Munz, Tiago M. Rodrigues, Jacek Krol, Tamas Szikra, Rachel Cuttat, Annick Waldt, Panagiotis Papasaikas, Roland Diggelmann, Claudia P. Patino-Alvarez, Patricia Galliker, Stefan E. Spirig, Dinko Pavlinic, Nadine Gerber-Hollbach, Sven Schuierer, Aldin Srdanovic, Marton Balogh, Riccardo Panero, Akos Kusnyerik, Arnold Szabo, Michael B. Stadler, Selim Orgül, Simone Picelli, Pascal W. Hasler, Andreas Hierlemann, Hendrik P. N. Scholl, Guglielmo Roma, Florian Nigsch, Botond Roska (2020)
Fast 3D Imaging of Spine, Dendritic, and Neuronal Assemblies in Behaving Animals. Gergely Szalay, Linda Judak, Gergely Katona, Katalin Ocsai, Gabor Juhasz, Mate Veress, Zoltan Szadai, Andras Feher, Tamas Tompa, Balazs Chiovini, Pal Maak, Balazs Rozsa, Neuron (2016)
Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules Wertz A, Trenholm S, Yonehara K, Hillier D, Raics Z, Leinweber M, Szalay G, Ghanem A, Keller G, Rozsa B, Conzelmann KK, Roska B, Science (2015)
Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes Gergely Katona, Gergely Szalay, Pál Maak, Attila Kaszas, Mate Veress, Daniel Hillier, Balazs Chiovini, E Sylvester Vizi, Botond Roska & Balazs Rozsa, Nature Methods (2012)
